NZOR Data Integration
NZOR Data Integration
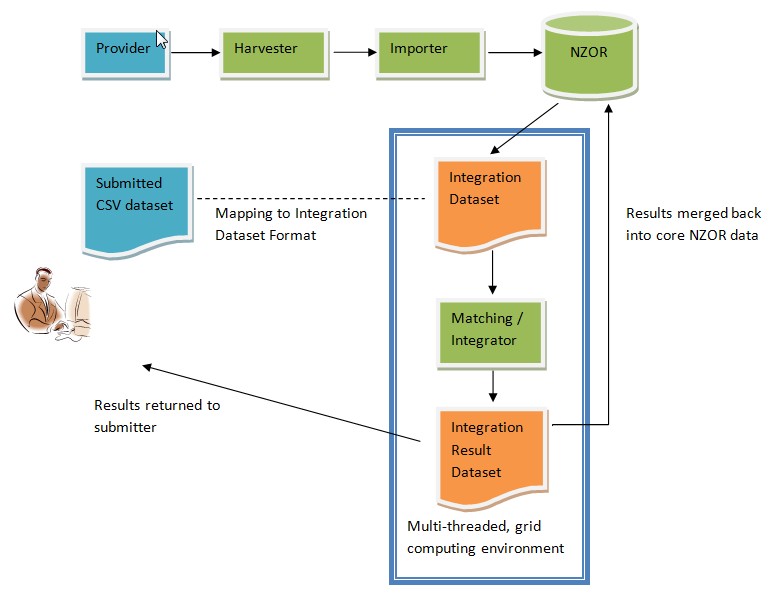
Workflow
UML Diagram
NZOR Integration Fields
2 types of matching – simple and structured. Simple is when only the Name text is provided and possibly other fields and Structured is when a set of detailed fields are provided.
For Simple the fields RequiredForMatching need to be derived/parsed/calculated from the fields that are provided.
Simple:
Field | Must be provided in provider dataset | Required for matching algorithm (must be provided or derived) |
FullName | true | false |
ProviderRecordId | false | true |
TaxonRank | false | true |
Authors | false | false |
GoverningCode | false | false |
ParentName | false | false |
PreferredName | false | false |
Structured:
Field | Must be provided in provider dataset | Required for matching algorithm (must be provided or derived) |
ProviderRecordId | true | true |
FullName | true | false |
TaxonRank | false | true |
GoverningCode | true | true |
Canonical | true | true |
ParentId | true | true |
Authors | false | false |
PreferredNameId | false | false |
YearOfPublication | false | false |
MicroReference | false | false |
PublishedIn | false | false |
Attachment Points
The highest ranked taxon name in a provider DataSet must either attach to a Kingdom name or a defined attachment point for that provider dataset.
Eg, a provider dataset may be the names for a particular family, say Compositae. An attachment point needs to be defined for this name (Compositae). This attachment point name MUST then be provided in the dataset so that it is possible to determine how to attach all subordinate names.
It is possible to have multiple attachment points, for different parts of the taxonomic classification hierarchy. There will need to be a Default attachment point defined for default actions such as placing a name that has unknown parentage.
Matching
Diagram:
Matching Components
Parent
The parent of a name defines where this name fits within a scientific classification, eg the Genus where a particular species is placed. (e.g. "Poa anceps" = the genus Poa is the parent name for the species "anceps"). This is a valuable property for the matching process.
Some types of names do not have a classification, e.g. Vernacular (common) names.
Name Type | Example | Has classification system |
Scientific | Poa anceps | Yes |
Vernacular (common) | Pohutakawa | No |
Trade name | Kiwifruit (??) | No (??) |
Tag name | "Gingidia | No (??) |
Multiple parent concepts
It is possible for a provider to maintain multiple parent concepts for a particular name, and therefore have that named placed under several different parents, depending on what authority they are following at the time (= multiple classifications). At this stage we are restricting data providers to supply a single "In Use" parent concept for a name (ie they can provide multiple parent concepts, but only one of those can be flagged as In Use). If there are multiple in use parent concepts or no in use parent concepts then further calculation is required. In this case all names that fall under any of the parents defined by the parent concepts are included in the matching. This will include a larger set of names to pass through to the next stage of matching, and if the matching algorithm concludes with more than one match due to the multiple parents, the match/integration will fail. If the algorithm concludes with no matches then, because of the multiple parent concepts, it is not possible to determine where to place this name. In this case the name is connected to the default attachment point for this dataset and should be flagged in some way to indicate this scenario.
How we handle this multiple classification situation into the future needs to be clarified.
Higher Level Names
It is possible for names at a higher level in the classification hierarchy (above genus) to be defined in different ways – different classifications. This should not prevent names at the same rank from matching even if their parent is not the same. For example a Family "Famae" is defined under the Order "Orderae" in one dataset and under the Order "Otherae" in another dataset. These names (Famae) are the same – i.e. they should match during integration. This means that the parent concepts defined by a particular dataset may often have to be overlooked during matching and integration.
Names with no parents
Trade names, tag names and vernacular names do not include a classification and therefore the integration must not include matching based on parent linkages.
Even scientific names with classification have no parent at the top of the hierarchy where this set of names needs to be attached to the NZOR dataset. This is determined by the attachment points defined in the admin for that particular provider.
There is a possible issue where the name as given by the submitter is at one rank and has a defined parent a rank above, but this is not reflected by the current NZOR data due to infraspecific ranks. For example the following name and parent name may be submitted for matching:
Name : Brickellia Elliott Rank : genus Parent : Compositae Rank : family
And the NZOR data could be:
Name : Brickellia Elliott Rank : genus Parent : Eupatorieae Cass. Rank : tribeGrandParent : Compositae Rank : family
And therefore filtering to all names with the parent "Compositae" will not find the NZOR genus "Brickellia". This issue is solved by filtering to all names with the Ancestor "Compositae" AND the Rank "genus".
Rank
The taxon rank is only applicable to scientific names. Vernacular names and trade names have a rank of "none". If the name that is being matched is a scientific name then the rank is used to filter the names down to those that are at the same rank as the name to be matched.
The rank provided by submitter must match one of the standard ranks in the NZOR system, or one of the "known abbreviations" of that rank.
For infraspecific names (i.e. those below the rank of species), the use of the rank is not covered by the codes of nomenclature and hence they are interchangeable, for example the infraspecific name Aus bus var. cus is equivalent to the name Aus bus subsp. cus. This must be taken into account during matching and integration.
Canonical
The canonical is the basic name text without any authors, year, publication details or parent information.
For example:
Brickellia Elliott Canonical : Brickellia Brickellia linifolia D.C.Eaton Canonical : linifolia
Craspedia "Henderson" Canonical : "Henderson"
Authors
Authors come in many forms and can be difficult to match against. One solution to this is to build a dictionary of standardised authors and their equivalents. Then when a submitted name is processed, the author is matched against the standardised list and the "standard" author name is then used to filter NZOR names for the match.
Data table structure for Author maintenance:
Field | Type |
AuthorId | Guid |
CorrectAuthorId | Guid |
Abbreviation | String |
FirstName | String |
LastName | String |
TaxonGroups | String |
Dates | String |
AlternativeNames | String |
Table for connecting a Name to its authors, basionym authors and combinations authors:
Field | Type | Description |
NameId | Guid |
|
BasionymAuthors | String | List of IDs of the Basionym Authors (non-standardised AuthorId value, space separated per author) |
CombinationAuthors | String | List of IDs of the Combination Authors (non-standardised AuthorId value, space separated per author) |
BasionymExAuthors | String | List of IDs of the Basionym "ex" Authors (non-standardised AuthorId value, space separated per author) |
CombinationExAuthors | String | List of IDs of the Basionym "ex" Authors (non-standardised AuthorId value, space separated per author) |
|
|
|
Year
Todo …
Nomenclatural Status
Some names have different nomenclatural status (e.g. whether the name was validly published under the code – if not the status = nom. inval.). It is possible for that name to be subsequently validly published. The details of the name will be the same, but the name now has a different nomenclatural status. At first it seems to be best to treat these as separate names that are linked. However this may cause issues during integration, for example if a provider does not provide the nomenclatural status, is this data for the validly published name or the invalid one? It was decided therefore to treat these names as the same and pool all nomenclatural status values for that name – it is then up to the consumer / viewer of that name to determine the status of the name. Therefore these names instances result in the same name, but there was two nomenclatural acts that led to this name.
Integration By SQL
A simpler, but much faster approach to building an initial set of integrated names is to use SQL queries.
The idea is based on the fact that most names are distinct (about 98%). It is therefore much more efficient to generate a "backbone" of names from these distinct names, rather than iterating through them all, performing a mathc to discover there are no matches, then inserting the name as a new consensus name.
This approach works with the most complete names first, with the theory that a name with less detail will match multiple names with more detail.
The fields that are used for defining a distinct name are:
- Canonical
- Rank
- Authors
- Year
- Genus
- Species
- GoverningCode
Genus and Species are not fields of a name, but are calculated fields based on parent concepts.The theory with including these fields is to ensure and sub generic name or sub specific name matches other sub generic/specific names that do not have exactly the same parent hierarchy.
For example:
Name 1: Aus bus var. cus
- Aus, genus
- bus , species
- cus, variety
- bus , species
Name 2: Aus bus xus var. cus
- Aus, genus
- bus, species
- xus, subspecies
- cus, variety
- xus, subspecies
- bus, species
The fields for these 2 names will be:
| Name | Canonical | Rank | Authors | Year | Genus | Species | Governing Code |
|---|---|---|---|---|---|---|---|
| 1 | cus | var. | Aus | bus | ICBN | ||
| 2 | cus | var. | Aus | bus | ICBN |
So according to these fields, the names will match even though the direct parents of the 2 'cus' names are different, which is correct.
Another example:
Name 1: Lecanorales Nannf., order
- Ascomycetes, class
- Lecanorales Nannf., order
Name 2: Lecanorales, order
- Ascomycetes, class
- Lecanoromycetidae, subclass
- Lecanorales, order
- Lecanoromycetidae, subclass
| Name | Canonical | Rank | Authors | Year | Genus | Species | Governing Code |
|---|---|---|---|---|---|---|---|
| 1 | Lecanorales | order | Nannf. | ICBN | |||
| 2 | Lecanorales | order | ICBN |
Again, will match even though the parent names are definied to be different. Again this is correct.
Generating Consensus Records
Order of save:
- Insert/update all References (as names and concepts rely on these)
- Update Provider References to point to the relevant Consensus Reference
- Refresh Consensus Reference data from all Provider records for modified references
- Insert update all Names (concepts rely on these)
- Update Provider Names to point to the relevant Consensus Name
- Refresh Consensus Name data from all Provider records for modified Names
- Insert/update all Concepts (not relationships as the relationships rely on both Concepts to be in existence)
- Update Provider Concepts to point to the relevant Consensus Concept
- Refresh Consensus Concept data from all Provider records for modified Concpets
- Insert/update all ConceptRelationships for each modified Concept, from all provider ConceptRelationship records
Technical Platform
Approaches:
- Direct connection to database, get next record to integrate, match and update the records in the database
- As in 1, but multithreaded (DB is the bottle neck)
- Load all data for integration into memory, then process the data in memory first, then save all changes to the DB (multi-threaded)
- As in 3, but run on a grid computing environment
Results:
- 0.8 records per second
- 1 record per second (threading not much advantage because DB is a bottleneck ??)
- 17 records per second
- ?
Conditions for generating another thread for the multi-threaded options include:
- Cannot integrate a name where the parent of that name has not been successfully integrated
- Cannot integrate a name that has any siblings that are currently being integrated
Possible improvements:
- Per thread, create 25 threads to process all provider names beginning with the same letter (as these are not likely to clash), or some other clustering approach – and perhaps only if there is > 100 child names ??
- Run a clustering algorithm over the provider names that need integrating before the integration is run to cluster unlikely matches (eg by first letter of name)
![worddav794473a6f96bfe5d4b0ddbeec67b688a.png [imported from a Word document]](https://manaakiwhenua.atlassian.net/wiki/download/thumbnails/4441047138/worddav794473a6f96bfe5d4b0ddbeec67b688a.png?version=1&modificationDate=1321980484840&cacheVersion=1&api=v2&width=1012&height=368)
![worddavd983a2c34317d761e05afaeba60c912e.png [imported from a Word document]](https://manaakiwhenua.atlassian.net/wiki/download/thumbnails/4441047138/worddavd983a2c34317d761e05afaeba60c912e.png?version=1&modificationDate=1321980485057&cacheVersion=1&api=v2&width=601&height=747)